One aspect of machine learning involves the development of generative models that are trained to generate variable output according to probabilistic information.
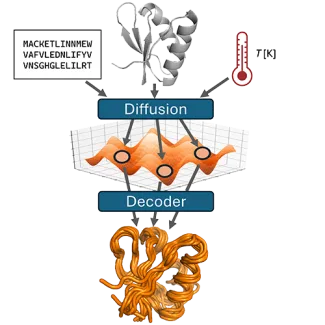
We are developing generative models for producing structural ensembles of biomolecules that reflect the dynamics of conformational sampling.
We are especially interested in training models based on molecular dynamics simulations to predict conformational ensembles for:
- intrinsically disordered peptides
- proteins around initial conformations
- conformational landscapes as a function of physical variables such as temperature
The initial goal is that such models will be able to replace traditional simulation methods at much reduced computational cost.
A longer-term goal is to capture biomolecular dynamics at experimental accuracy beyond limitations from using MD simulation data for training.
Software and modeling resources: