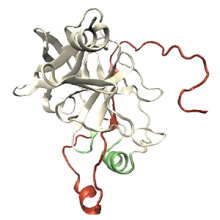
Accurate Predictions of Molecular Properties of Proteins via Graph Neural Networks and Transfer Learning
J. Chem. Theory Comput. (2025)
Controlled Enzyme Cargo Loading in Engineered Bacterial Microcompartment Shells
Biochemistry (2025) 64, 1285-1292
Structure Characterization of Bacterial Microcompartment Shells via X-ray Scattering and Coordinate Modeling: Evidence for Adventitious Capture of Cytoplasmic Proteins
ACS Appl. Bio Mater. (2025) 8, 2090-2103
COCOMO2: A Coarse-Grained Model for Interacting Folded and Disordered Proteins
J. Chem. Theory Comput. (2025), 21, 2095–2107
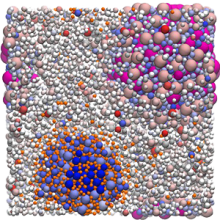
Diffusion and Viscosity in Mixed Protein Solutions
J. Phys. Chem. B (2024), 128, 11676–11693
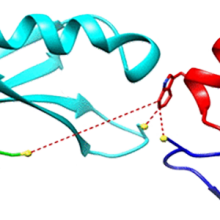
De Novo Synthesis and Structural Elucidation of CDR-H3 Loop Mimics
ACS Chem. Biol. (2024), 19, 1583–1592
Stability and deformation of biomolecular condensates under the action of shear flow
J. Chem. Phys. (2024),160, 215101
Transferable deep generative modeling of intrinsically disordered protein conformations
PLoS Comp. Biol. (2024), 20, e1012144
One bead per residue can describe all-atom protein structures
Structure (2024), 32, 97-111.e6
The effect of polymer length in liquid-liquid phase separation
Cell Reports Physical Science (2023), 4, 101415
Direct generation of protein conformational ensembles via machine learning
Nat. Comm. (2023) 14, 774
Varying molecular interactions explain aspects of crowder-dependent enzyme function of a viral protease
PLoS Comput. Biol. (2023), 19, e1011054
Characterization of RNA polymerase II trigger loop mutations using molecular dynamics simulations and machine learning
PLoS Comput. Biol. (2023), 19, e1010999
Modeling Concentration-dependent Phase Separation Processes Involving Peptides and RNA via Residue-Based Coarse-Graining
J. Chem. Theory Comput. (2023), 19, 669-678
Molecular Dynamics Simulations of Rhodamine B Zwitterion Diffusion in Polyelectrolyte Solutions
J. Phys. Chem. B (2022), 126, 10256-10272
Multi-state modeling of G-protein coupled receptors at experimental accuracy
Proteins (2022), 90, 1873-1885
Characterizing Transient Protein-Protein Interactions by Trp-Cys Quenching and Computer Simulation
J. Phys. Chem. Lett. (2022), 13, 10175-10182
Crowding affects structural dynamics and contributes to membrane association of the NS3/4A complex
Biophys J. (2021), 120, 3795-3806
Charge-driven condensation of RNA and proteins suggests broad role of phase separation in cytoplasmic environments
eLife (2021), 10, e64004
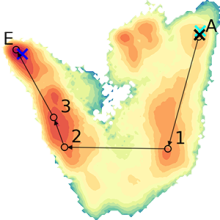
Improved Sampling Strategies for Protein Model Refinement Based on Molecular Dynamics Simulation
J. Chem. Theory Comput. (2021), 17, 1931–1943
Physics-based protein structure refinement in the era of artificial intelligence
Proteins (2021), 89, 1870-1887
Reduced efficacy of a Src kinase inhibitor in crowded protein solution
Nat. Comm. (2021), 12, 4099