Computational studies of biomolecular systems are fundamentally challenging. In order to make meaningful predictions and connect with experiments it is necessary to:
- Achieve sufficient physical accuracy
- Reach large enough system sizes to avoid finite-size effects
- Cover long-enough time scales to match experiments and reach biological relevance
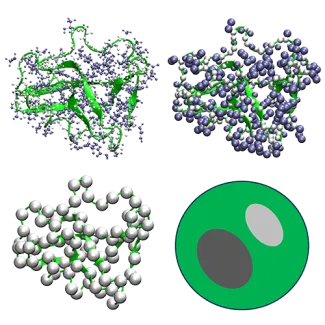
Multiscale modeling is a strategy for describing systems simultaneously at different scales to achieve both accuracy and the ability to reach large system sizes and long time scales.
We have developed coarse-grained models at different resolutions that can connect directly with experiments and biology. Models developed by us include the high-resolution PRIMO model, the residue-level COCOMO model, and sphere models of biomolecules.
A key challenge is the connection between different levels. Most recently we have proposed the cg2all method for mapping between coarse-grained and atomistic models of proteins.
Software and modeling resources:
- MMTSB Tool Set: tools for multiscale modeling
- COCOMO: residue-level coarse-graining of proteins and RNA
- cg2all: reconstruction of atomistic detail from coarse-grained representations